
Whole genome sequencing is a complete process for analysing the whole genomes of any data. This technique has transformed the field of genomics and has several applications in various fields, including biotechnology, agriculture, and healthcare. One of the major advantages of WGS is that it gives a complete view of an organism genetic makeup, allowing researchers to study for complete set of genes, including those that are not well-categorised. This information used to identify variation in genes that may be associated with the diseases and can help to develop more actual treatments for a variation.
Generally, WGS is an important tool’s for progressing scientific understanding and improving human and environmental health. By providing a over all view of an organism’s genetic makeup, WGS has the possible to enable a wide range of applications, from personalized medicine to the development of more tough crops and livestock. As per technology and tools to improve and become more accessible, it is likely that we will see even more innovative uses for WGS in the years to come.
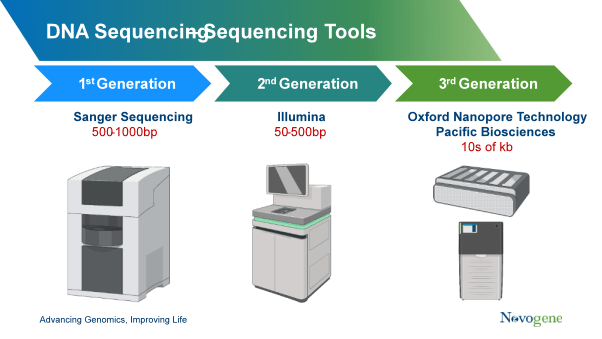
WGS is the most universal approach to identifying genetic differences. Though in humans sample the first genome analysis was used in sanger sequencing and for this process cost is high and it take more time as compared to other genomic sequencing methods. For this process some of biotechnology company like illumina sells multiple next generation sequencing platform where whole genome sequencing is analysed easily and take less than of 30 hours.
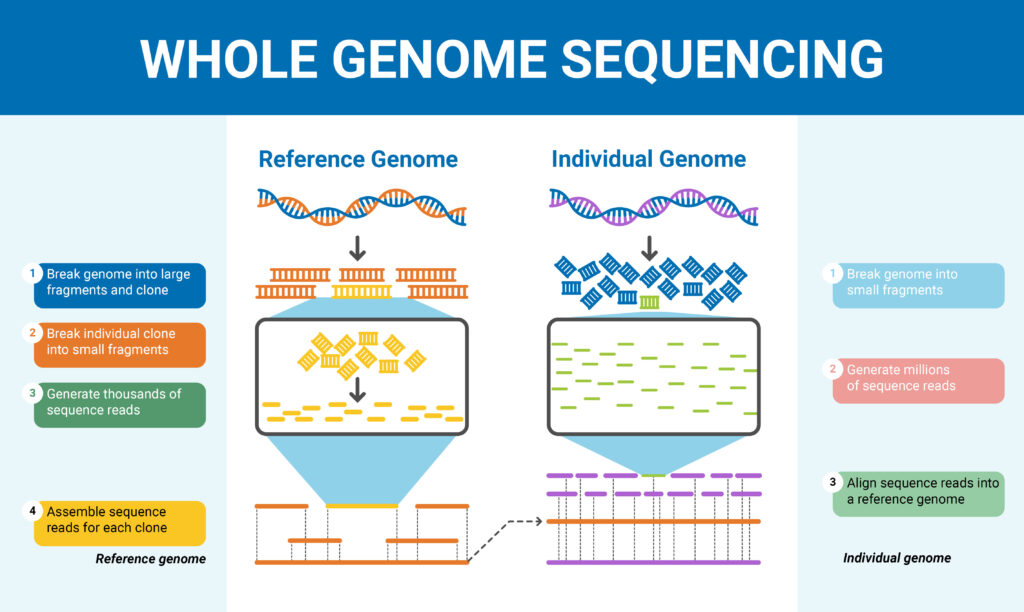
Whole genome sequencing can find out the all large structural variants, insertions or deletions, copy number changes, and single nucleotide variants. Due to recent innovations, the novel genome sequencers can perform whole genome sequencing more efficiently as compared to old approaches.
Emerging Applications and Advances in Whole Genome Sequencing
Large Whole Genome Sequencing
Sequencing large genomes such as human, animal and plant genomes, it can provide actual information for disease research and population of genetics.
DeNovo Sequencing
It refers to sequencing of a novel genome where there is no reference genome sequence available. NGS enables it fast and accurate the characterization of any species.
Human Whole Genome Sequencing
Most detailed view into our genetic code
Small Whole Genome Sequencing
Small genome sequencing involves sequencing the entire genome of a virus, bacterium or other microbes. Without requiring bacterial culture, researchers can sequence the thousands of small organisms in a parallel sequencing using NGS.
Phased Sequencing
Phased sequencing or genome phasing it differentiates between alleles on homologous chromosomes, resulting in whole genome haplotypes. This information is important for genetic disease studies.
Long Read Sequencing
It can help in resolve challenging regions in the genome, such as those containing highly variable or highly repetitive elements.
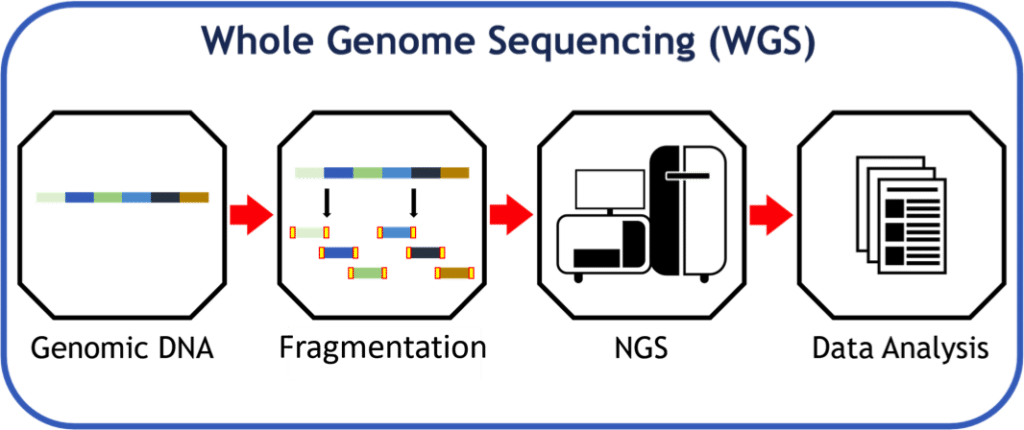
Steps for whole genome sequencing
- Data preparation
- Sample extraction and purification
- Library construction
- Whole sequencing implementation
- Alignment and assembly
- Map the sequence reads to reference sequence
- Reconstruct the original sequence
- Quality control
- Variant calling
- Single nucleotide variant calling
- Indels calling
- Structural and copy number variant calling
- Annotation and analysis
- Filtering and annotation of matching segment
- Detection of crucial variant related to disease
- Variant analysis to the development of pathogen
Platform for WGS work analysis
- Illumina
- Galaxy
- Pacific-bioscience
- Oxford nanopore
- LONI
- GATK
- Genestack
- triana
Application of whole genome sequencing
- Personalized medicine
- Disease causing mutation
- Application into agriculture
- Used in drug discovery
- Cancer biology
- Forensic science
TOOLS USED IN WGS
- PREPROCESSING
- Sbs
- Smart
- Swift
- Short Read
- Fqc
- ALIGNMENT
- Zoom
- Bowtie
- Bwa
- ASSAMBLY
- Valvet
- Soa Pdenovo
- Maq
- Eland
- Rays
- SNV/INDELS
- Samtools
- Gatk
- SVs/CNVs
- Parliament
- Delly2
- ANNOTATION
- Annovar
- Vat
- Snpeef
Steps and tools involve in data analysis in whole genome sequencing using NGS
| Steps | Tools | descriptions |
| RAW READ | ||
| Quality analysis | fastqc | Quality checkup of raw sequence data |
| Trimming the Bad Quality Reads | Trimmomatic | Cut adopter and other illumina specific seq from the reads |
| Quality Analysis – Trimmed Reads | fastqc | Quality checkup of trimmed data |
| Mapping of Trimmed Raw Reads with Reference Genome | MAP with BWA | Mapping include low divergent sequence against a large reference genome which is mm10, BWA design for illumine seq up to for 100 bp. |
| Removal of Unmapped Reads | Filter SAM or BAM samtools flagstat rmdup | Filter a SAM or BAM file on mapping quality .Print descriptive information for a BAM dataRemove potential pcr duplicates |
| Variant Mapping | Free bayes | Variant analysis find small polymorphism, SNP(single nucleotide polymorphism),MNP(multi nucleotide polymorphism),insertion and deletions. |
| Variant of Interest | SNPEFF download ,snpeff eff |
Advantages of Whole Genome Sequencing using Bioinformatics
- Provides a high-determination, base-by-base visualisation of the genome
- Analysed both small and large variants that missed with targeted approaches
- Identifies likely causal variants for more follow up studies of gene regulation and expression mechanisms of the gene.
- Delivers a large amount of data in a short interval of time to support assembly of novel genomes
Conclusion
With the help of bioinformatics tool and techniques biological science became easy as compared to old approaches and techniques. whole genome analysis is the process where whole genome sequencing comes out with computational bioinformatics . For any biological process bioinformatics take less time and the cost is very less as compared to others approaches. For medical purpose our body is made up of millions of cell and our body contain genetic material such as DNA , RNA and protein.
From this genetic material with the help of next generation sequencing we can sequence protein coding region exon and intron easily Each gene are divided in to exons and introns and exons are involved in making proteins most of the mutations occur in exons regions from this changes Autosomal dominant disorder and autosomal recessive disorder caused in specific gene. Genome sequencing help out for this type of disorder. For this type of disease doctors suggest the genome sequencing data analysis for any variations in the cells.
On the basis of analysis of data with the help of next generation sequencing the final analysis creates a result on the basis of graphs, data and compare that results with other approaches easily.
To know more about the Whole Genome Sequencing process or to understand how the process works you can join us for a 3 Hours Short Course on Genome Analysis, you can register yourself HERE
In this article we discussed about the analysis of DNA data but if you want to know about analysis of RNA data you can refer to the article HERE and if you are interested in understanding it’s application in detail you can find it HERE
This article is originally published HERE







0 Comments