
In the late 1990s, geneticist Dr. Jane Goodall joined the Human Genome Project, driven by her family’s history of a rare genetic disorder. Her meticulous work led to the discovery of a single nucleotide polymorphism (SNP) linked to the disorder, a breakthrough that enabled early diagnosis and potential treatments. This finding not only offered hope to her family but also highlighted the power of genome analysis in understanding genetic variations and their health impacts. Today, genome analysis continues to revolutionize medicine and biology, uncovering secrets and transforming lives.
Genome analysis is a powerful and rapidly advancing field that involves examining the structure, function, and evolution of genomes—the complete set of DNA within an organism. The advancements in this area have revolutionized our understanding of biology and medicine, enabling researchers to uncover the genetic basis of diseases, trace evolutionary histories, and develop personalized medical treatments.
At the core of genome analysis is the sequencing of DNA, which determines the exact order of the nucleotides (adenine, thymine, cytosine, and guanine) in a genome. The development of high-throughput sequencing technologies has dramatically reduced the cost and time required for genome sequencing.
Methods such as next-generation sequencing (NGS) allow for the parallel sequencing of millions of DNA fragments, making it possible to sequence entire genomes quickly and accurately. These advancements have made genome analysis accessible to a wide range of researchers and have opened up new possibilities for genetic research.
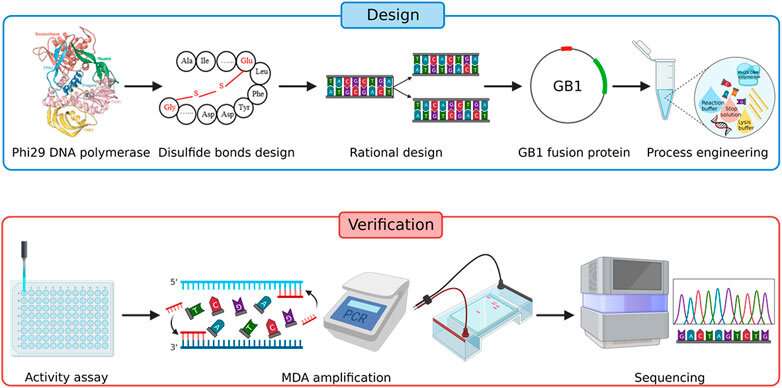
One of the primary applications of genome analysis is in the field of medical genetics. By comparing the genomes of individuals with and without certain diseases, researchers can identify genetic variants associated with those conditions. This approach has led to the discovery of numerous disease-associated genes and has provided insights into the genetic underpinnings of complex diseases such as cancer, diabetes, and heart disease.
Furthermore, genome analysis has paved the way for the development of personalized medicine, where treatments can be tailored to an individual’s genetic makeup. For example, certain cancer therapies are now chosen based on the genetic mutations present in a patient’s tumor, increasing the effectiveness of treatment and minimizing side effects.
Another significant application of genome analysis is in evolutionary biology. By comparing the genomes of different species, scientists can reconstruct the evolutionary relationships between them and trace the origins and migrations of populations. This has provided a deeper understanding of the tree of life and has helped to uncover the genetic basis of adaptation and speciation.
For example, genome analysis has revealed how certain species have adapted to extreme environments, such as high altitudes or deep-sea habitats, by identifying specific genetic changes that confer survival advantages.
In addition to its applications in medicine and evolutionary biology, genome analysis is also used in agriculture and conservation biology. In agriculture, the sequencing of crop and livestock genomes has enabled the identification of genes associated with desirable traits, such as disease resistance, yield, and nutritional content.
This information is being used to develop genetically modified organisms (GMOs) and to guide traditional breeding programs, leading to the production of more resilient and productive crops and livestock. In conservation biology, genome analysis is used to assess the genetic diversity of endangered species, identify populations at risk of inbreeding, and develop strategies for their conservation and management.
The field of genome analysis also encompasses the study of the epigenome and transcriptome. The epigenome consists of chemical modifications to the DNA and associated proteins that regulate gene expression without altering the DNA sequence. These modifications can be influenced by environmental factors and can be inherited across generations, playing a crucial role in development and disease.
Genome analysis techniques, such as bisulfite sequencing, are used to map these epigenetic modifications and to understand their impact on gene function. The transcriptome, on the other hand, represents the complete set of RNA molecules transcribed from the genome, providing a snapshot of gene expression at a given time. RNA sequencing (RNA-seq) is a powerful tool for transcriptome analysis, allowing researchers to quantify gene expression levels, identify alternative splicing events, and discover novel transcripts.
One of the challenges in genome analysis is the interpretation of the vast amount of data generated by sequencing technologies. Bioinformatics plays a crucial role in this process, providing the tools and algorithms needed to analyze and interpret genomic data. This involves aligning sequencing reads to a reference genome, identifying genetic variants, and annotating genes and regulatory elements.
Advanced computational methods, such as machine learning and artificial intelligence, are increasingly being used to identify patterns in genomic data and to predict the functional consequences of genetic variants. The integration of genomic data with other types of biological data, such as proteomics and metabolomics, is also becoming more common, providing a more comprehensive view of the molecular mechanisms underlying biological processes.
The ethical, legal, and social implications of genome analysis are also important considerations. The ability to sequence and analyze genomes raises questions about privacy, consent, and the potential for genetic discrimination. The storage and sharing of genomic data must be carefully managed to protect individuals’ privacy while enabling scientific research.
Furthermore, the use of genome editing technologies, such as CRISPR-Cas9, which allow for precise modifications to the genome, has sparked debates about the ethical implications of altering human DNA. The development of guidelines and regulations to address these issues is essential to ensure the responsible use of genome analysis technologies.
As the field continues to advance, the integration of genomic data with other types of biological data and the development of new computational methods will further enhance our understanding of the complex mechanisms underlying life. However, the ethical, legal, and social implications of genome analysis must be carefully considered to ensure the responsible and equitable use of these powerful technologies.
What is Genome Analysis
Genome analysis is a pivotal scientific field that involves studying the complete set of DNA within an organism, known as its genome. This comprehensive analysis helps researchers understand the structure, function, and evolution of genes, providing crucial insights into genetic diseases, evolutionary biology, and personalized medicine.
But What is genome analysis? At its core, genome analysis involves sequencing, which determines the exact order of the nucleotides in DNA. Advances in high-throughput sequencing technologies have made it possible to rapidly and accurately sequence entire genomes. By comparing genomic sequences from different individuals or species, scientists can identify genetic variations associated with diseases, trace evolutionary relationships, and understand genetic adaptations.
Then What does genome mean? A genome is the complete set of an organism’s genetic material, encompassing all its genes and non-coding sequences. It is essentially the blueprint for building and maintaining that organism. Genome analysis enables researchers to decode this blueprint, leading to groundbreaking discoveries in various fields, from medical genetics to agriculture.
In medicine, genome analysis has led to the identification of numerous disease-causing genes, paving the way for targeted therapies and personalized treatments. In agriculture, it aids in the development of crops with desirable traits, such as disease resistance and improved yield. Overall, genome analysis is transforming our understanding of biology and driving innovations across multiple domains.
Pan Genome Analysis
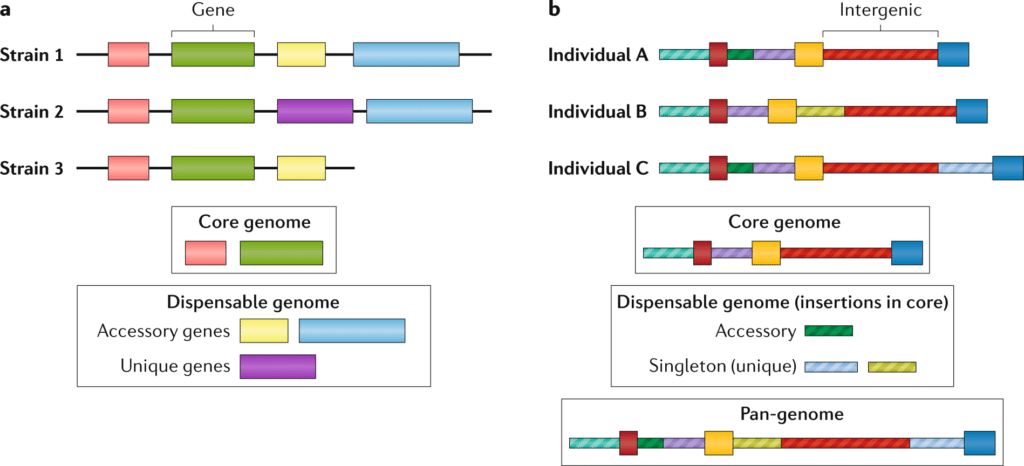
Pan genome analysis is a cutting-edge technique used to study the complete set of genes within a species, encompassing the core genome shared by all individuals and the accessory genome that includes variable genes. This comprehensive approach provides a deeper understanding of genetic diversity, evolution, and adaptation within a species, revealing insights that are critical for fields such as microbiology, agriculture, and medicine.
What technique is used to analyze genomes in pan genome analysis? High-throughput sequencing technologies, such as next-generation sequencing (NGS), are pivotal. These methods allow for the rapid and accurate sequencing of multiple genomes from different strains or individuals within a species. By comparing these sequences, researchers can identify the core genes essential for survival and the accessory genes responsible for unique traits and adaptations.
Pan genome analysis is particularly valuable in studying bacteria and plants. In microbiology, it helps to understand pathogenicity, antibiotic resistance, and microbial evolution. In agriculture, it aids in breeding programs by identifying genes associated with disease resistance, yield, and other desirable traits.
Overall, pan genome analysis offers a comprehensive view of the genetic landscape within a species, enabling scientists to uncover the complexities of genetic variation and apply this knowledge to various scientific and practical applications.
Genome Analysis Methods
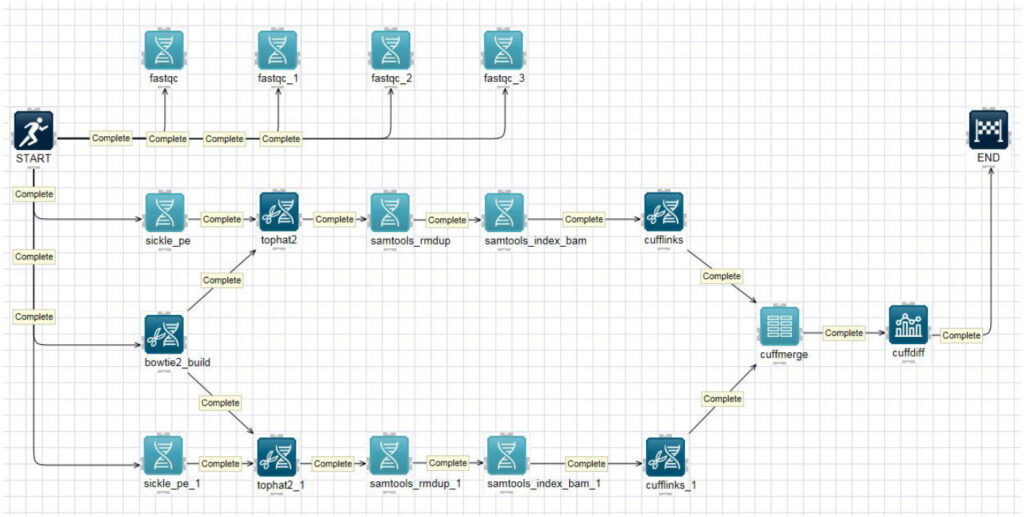
Genome analysis methods are a suite of techniques used to decode and understand the complete set of DNA within an organism. These methods are crucial for unraveling the genetic blueprint of organisms, leading to significant advancements in medicine, agriculture, and evolutionary biology.
The foundation of genome analysis methods lies in DNA sequencing, particularly next-generation sequencing (NGS). NGS allows for the rapid and high-throughput sequencing of entire genomes, providing detailed information on genetic variations and mutations. Whole-genome sequencing (WGS) is a comprehensive method that sequences an organism’s entire genome, offering a complete genetic picture.
Another important method is exome sequencing, which focuses on the coding regions of the genome that directly translate into proteins. This approach is cost-effective and useful for identifying mutations that cause genetic disorders. Additionally, RNA sequencing (RNA-seq) analyzes the transcriptome, revealing which genes are actively expressed and how they are regulated.
Genome-wide association studies (GWAS) are employed to identify genetic variations linked to specific diseases or traits by scanning the genomes of many individuals. Comparative genomics involves comparing genomes across different species to understand evolutionary relationships and functional genomics.
These genome analysis methods have revolutionized our understanding of genetics, enabling personalized medicine, enhancing crop yields, and providing insights into the complexities of life. As technology advances, these methods continue to evolve, offering even deeper insights into the genetic underpinnings of organisms.
Genome Analysis Tools
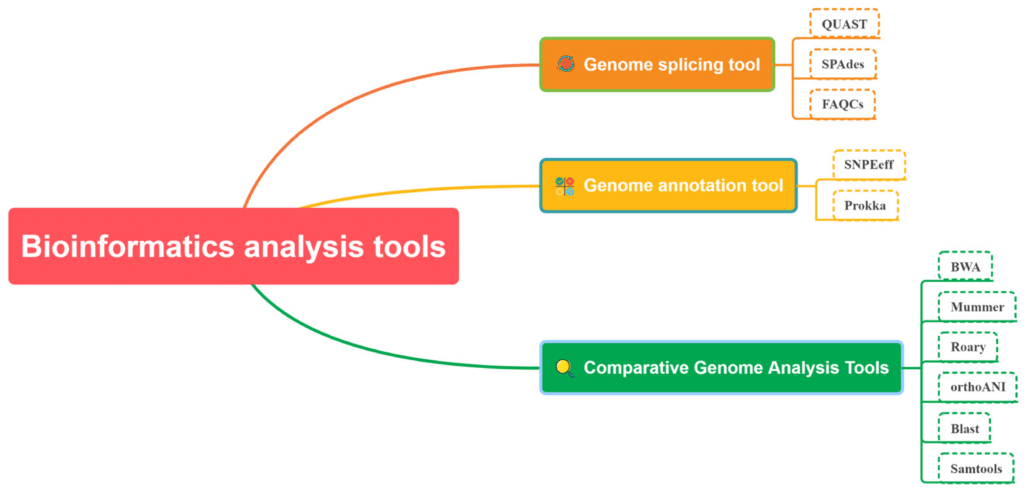
Genome analysis tools are essential software and computational resources used to interpret the vast amount of data generated by DNA sequencing. These tools enable researchers to decode, analyze, and understand the genetic information contained within genomes, facilitating advancements in medicine, agriculture, and biological research.
One of the primary genome analysis tools is the Basic Local Alignment Search Tool (BLAST). BLAST allows researchers to compare an unknown DNA sequence against a database of known sequences, identifying similarities and potential functions. Another widely used tool is the Genome Analysis Toolkit (GATK), which is instrumental in identifying and annotating genetic variants. GATK’s robust algorithms ensure high accuracy in variant calling, making it a cornerstone in genetic research.
For visualizing genomic data, Integrative Genomics Viewer (IGV) is a powerful tool. IGV provides an interactive interface to explore and visualize large genomic datasets, helping researchers identify patterns and anomalies. Additionally, bioinformatics platforms like Galaxy offer user-friendly interfaces for performing complex genomic analyses without requiring extensive programming knowledge.
Tools like PLINK are used for genome-wide association studies (GWAS), enabling the identification of genetic variants associated with specific traits or diseases. For RNA sequencing analysis, tools like STAR and HISAT2 facilitate efficient alignment of RNA sequences to reference genomes.
Overall, genome analysis tools are indispensable in converting raw sequencing data into meaningful biological insights, driving progress across various scientific fields. As technology advances, these tools continue to evolve, offering enhanced capabilities and greater accuracy in genomic research.
Comparative Genome Analysis

Comparative genome analysis tools are essential for understanding the genetic differences and similarities between different species or individuals within a species. These tools facilitate the comparison of entire genomes, providing insights into evolutionary relationships, functional genomics, and the genetic basis of phenotypic traits.
One key comparative genome analysis tool is Mauve, which aligns multiple genomes to identify regions of similarity and difference. Mauve is particularly useful for studying evolutionary changes and rearrangements in bacterial genomes. Another important tool is OrthoMCL, which clusters orthologous genes across multiple genomes, helping researchers identify conserved gene functions and evolutionary relationships.
The Ensembl Genome Browser is a comprehensive resource that provides access to genomic data from numerous species, allowing researchers to compare genomic regions and gene structures. Ensembl’s integrated tools, such as the Comparative Genomics module, enable detailed analysis of synteny, orthology, and gene conservation.
For large-scale genomic comparisons, tools like Panseq are invaluable. Panseq analyzes the pan-genome of a species, identifying core and accessory genes that contribute to genetic diversity. Another powerful tool, SyMAP, generates synteny maps that visualize genomic alignments, making it easier to identify conserved genomic regions and evolutionary events.
Overall, comparative genome analysis tools are crucial for elucidating the complexities of genetic evolution and diversity. These tools provide the necessary infrastructure to decode evolutionary processes, understand genetic variations, and apply this knowledge to various biological and medical fields. As genomic data continues to grow, these tools will become even more sophisticated, offering deeper insights into the genetic fabric of life.
Main Function & Role of Genome Analysis
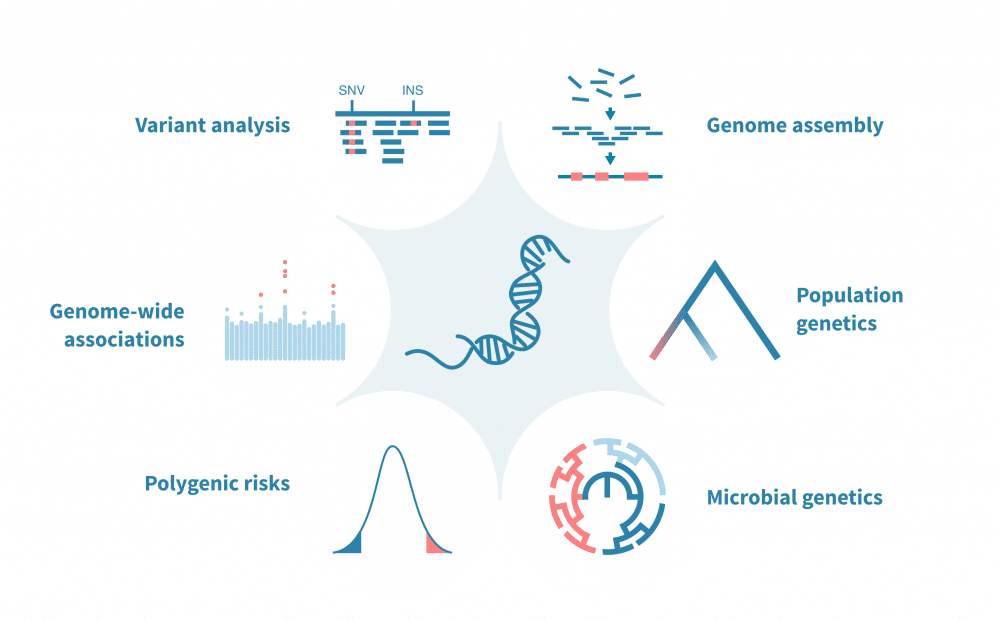
The main function and role of genome analysis tools are pivotal in the field of genomics, where they facilitate the interpretation and understanding of the vast amounts of genetic data generated by DNA sequencing. These tools are crucial for decoding the genetic blueprint of organisms, enabling advancements in medicine, agriculture, and evolutionary biology.
One of the primary functions of genome analysis tools is to sequence and assemble genomes. Tools like the Genome Analysis Toolkit (GATK) and SPAdes play essential roles in ensuring accurate and efficient genome assembly, allowing researchers to construct complete genetic maps. These assembled genomes provide the foundation for further analysis and research.
Genome analysis tools are also vital for identifying genetic variants. Software such as GATK and SAMtools are used to detect single nucleotide polymorphisms (SNPs) and insertions/deletions (indels) within the genome. Identifying these variants is crucial for understanding genetic diversity, disease susceptibility, and evolutionary adaptations.
Functional annotation is another key role of genome analysis tools. Tools like BLAST and Ensembl help annotate genes and predict their functions by comparing them to known sequences. This information is essential for understanding gene regulation, expression, and interactions.
Moreover, comparative genome analysis tools, such as Mauve and OrthoMCL, allow researchers to compare genomes across different species or individuals. This comparative approach is fundamental for studying evolutionary relationships, identifying conserved genetic elements, and understanding the genetic basis of phenotypic traits.
Overall, the main function and role of genome analysis tools are to transform raw genetic data into meaningful biological insights. They are indispensable for advancing our understanding of genetics and driving innovations in various scientific and practical applications.
In Conclusion, Genome analysis is essential for decoding genetic blueprints, enabling breakthroughs in medicine, agriculture, and evolutionary biology. Through advanced sequencing and comparative tools, it uncovers genetic variations, evolutionary relationships, and functional annotations, transforming vast genetic data into actionable insights that drive scientific and practical innovations across multiple fields.
To Know More about the Functioning and Analysis Pipeline of Genome Analysis Please Feel Free to Join us for a 3 Hour Short Course, You can register HERE for the same.
You can ready About the 4 Key Applications of Next Generation Sequencing Data Analysis HERE, a brief guide is available.







0 Comments